Users' documentation for BMapBuilder
Installation
Java Runtime Environment
BMapBuilder is a Java 2 program. In order to use it, you need a Java Runtime Environment (JRE) version 1.4 or later. If you do not have it, get and install the newest version of the JRE at:
Sun JRE
Java 3D
Since version 2 of BMapBuilder, 3D functionality was added. For 3D functions to be run in your computer you will need to have installed Java 3D, at least version 1.3.1:
Download Java3D.
If you perform a manual installation, please make sure files are installed in the correct folder or the program will not start.
Incompatibility notes
It has to be noted that recent versions of Java3D may not be supported by some graphic drivers. If 3D functionality of BMapBuilder 2.0 does not work, you may need either to uninstall Java3D and install and older version (we recommend version 1.3.1 or 1.3.2) or to update your graphic driver.
Download BMapBuilder
Download BMapBuilder 3.2 with 3D functionality (recommended)
Download BMapBuilder 3.1 with 3D functionality
Download BMapBuilder 2.3 with 3D functionality
Download BMapBuilder 2.2 with 3D functionality
Download BMapBuilder 2.1 with 3D functionality
Follow version control
Install BMapBuilder
Windows, Mac OSX
Make sure that the file you have downloaded has extension ".jar", as some Internet browser may change it by ".zip" when downloading. You should be able to start the program by double-clicking on the jar file.
The program can also be run by typing java -jar [BMapBuilder jar file] on the command line.
Unix, Linux
The program can be run by typing java -jar BMapBuilder2.jar on the command line or by double-clicking on it.
Large datasets
We recommend to use the argument -Xmx for large datasets. Otherwise the program may return an "out of memory" error.
Thus, for large datasets, as chromosome-wide datasets, the program must be run by typing java -jar -XmxMm BMapBuilder2.jar, with M being the maximum internal memory your computer can use in Megabytes. As an example,
java -jar -Xmx2000m BMapBuilder3.1.jar
will allow the program to use a maximum of 2000 MB memory.
Textual user interface
For large data files we strongly recommend to use a textual user interface, as the following example:
java -jar -Xmx2000m BMapBuilder3.1.jar -ng -f=i.pm -o=o.png -est=8 -snp1=1 -snp2=4
.
-Xmx2000m will allow the program to use a maximum of 2000 MB memory
-ng will run the program in textual mode
-f=i.pm will use file i.pm as the input file
-f=o.png will use file o.png as the output file
-est=8 will use information at column 8 as LD measure
-snp1=1 will use column 1 to find the position of SNP 1
-snp2=4 will use column 4 to find the position of SNP 2
Run java -jar BMapBuilder -ng for more information about program arguments.
Input file format
Text files with at least three columns, no matters the order, are required.
Each row contains information for a pair of SNPs.
The three columns correspond to the physical position of each SNP in the pair and the estimation value (which must be a value in the interval [0,1]).
Besides these three columns, the program can also uses the major allele relative frequency for each SNP in the pair, i.e., two more columns.
The order is not important as the program allows the user to choose the colums.
Other columns in the file will not be taken into account.
Here there is a very small file than can be used as an example of an input file.
There can be a first line in the row with the heading. The number of words in the heading must be the same as columns in the file. Words must be separated by tab, blank space or comma.
Output file format
The bitmap will be written in a png graphic file.
For each snp pair, a square with res bits side will be used to represent the value of the estimation, following this scale:
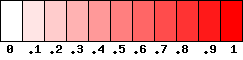
The value of res can be chosen in the program as well as the color tones (green, red or blue).
We do not recommend resolutions greater than 5 for large files, as those at the International HapMap Project website.
How to use BMapBuilder
In order to build a new 2D/3D bitmap using the Graphical User Inteface (GUI), you have to use the option "Build" in the "2D/3D" menu.
Besides providing a source file, the following mandatory parameters will be required to the user when a 2D bitmap is going to be built, either from the GUI or from the command line:
- Column where position for the first SNP is
- Column where position for the second SNP is
- Column where the estimator (D', r, LOD or whatever) is
The following parameters are optional:
- Whether the source file contains a heading line (no by default)
- Column with a confidence level if the map is to be coloured using also the confidence level of the LD measure estimation
- Binary signal column.- It is used to represent a single individual versus sample recombination LD maps. 0 means 2-snp haplotype is unobserved, 1 means it is observed and consistent with the non recombinant haplotypes (it will be represented in green color for maps in red or blue, blue for maps in green) and -1 means it is observed and it is consistent with recombination haplotypes (blue color for maps in red, red color for maps in green or blue).
- SNP1 column for annotation.- Column number with annotations used for SNP1 instead of that at the SNP1 column
- SNP2 column annotation.- Column number with annotations used for SNP2 instead of that at the SNP2 colums.
- Annotation.- A file with the SNP positions to be shown in the LD map. If not used, SNP positions will be shown in the LD map at a constant distance.
- Annnotation 2.- A second file with SNP positions to be shown in green color (instead of black). If a position is also included in the "Annotation" file, it eill be shown in red color.
- Individual pos.- Optional field for individual versus sample recombination LD map (minimum size for each LD pair (in the following menu if using the GUI) must be set to 3): choose a file with individual positions to be shown in the LD map. color black is used whether individual sets are inconsistent between a pair of adjacent squares, white if they are consistent.
- MAF1.- MAF for the first SNP in the case a threshold is to be imposed to display the first SNP.
- MAF2.- MAF for the second SNP in the case a threshold is to be imposed to display the second SNP.
- Color to be used (red, green or blue).- If using the GUI, this parameter will be entered in a second window.
- Resolution to be used (for larga data sets, high resolutions should only be used if the computer has enough RAM memory).- If using the GUI, this paramenter will be entered in a second window.
Citation
Please, cite BMapBuilder with the following paper:
Abad-Grau, M.M.; Montes, R.; Sebastiani, P.
"Building chromosome-wide LD maps",
Bioinformatics 22(16): 1933-1934 (2006)
Additional information
Some datasets
Other supplementary information
